Understanding the proteins language:
Is the future medicine here?
Tamam Abdul-Ghani# and Mickey Kosloff - Department of Human Biology
Rachel Kolodny - Department of Computer Science
Machine Learning
Deep learning
Neural Networks
Health
PhD Grant 2021
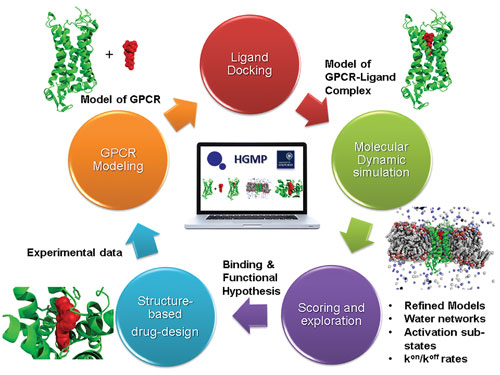
The interaction between proteins in our body cells is the biological language that determines our health and illness. Cellular functions are regulated by a plethora of extracellular signals such as sensory stimulus, hormones, or neurotransmitters. Many of these signals act through receptors on the cell surface to modulate intracellular signaling cascades. G protein coupled receptors (GPCRs) compose an essential proteins family that responsible for signaling between our cells and organs (Pierce et al., 2002). GPCRs modulate many physiological processes and are responsible for a wide range of diseases, with about 40% of marketed drugs acting on these signaling pathways (Santos et al., 2017). Human GPCRs derive from approximately 800 different genes that are classified into six classes (A-F) based on structural and functional similarity (Lagerström et al., 2008).
Over the last decade, many hundreds of GPCR 3D structures with ligands or protein partners were determined, shedding light on the extracellular and intracellular interactions of individual GPCRs. For example, dozens of 3D structures of the glucagon-like peptide 1 receptor and related GPCRs have been determined in recent years, leading to a better understanding of how this receptor modulates metabolism and regulates diabetes (Cho, Merchant and Kieffer, 2012). This information has enabled the design of new drugs for diabetes that are already on the market, but these drugs have side effects and their efficacy can be much improved.
However, the lack of computational tools to analyze and compare the vast amount of structural data available and combine them with orthogonal information such as sequences limits the ability of scientists to extract broad yet precise data that will enable to maximize the scientific and therapeutic potential of GPCRs. The methods we intend to develop in our research will provide the ability to analyze numerous 3D structures precisely. In addition, computational approaches that better utilize the vast amount of 3D structural data for GPCR families can be the basis for developing future approaches for deciphering the interactions among other large protein families.

